By: John Miner | Updated: 2023-03-21 | Comments (1) | Related: > Apache Spark
Problem
Extensible Markup Language (XML) is a markup language and file format. It is commonly used in web applications for storing, transmitting, and reconstructing arbitrary data. This transfer technique has been referred to as the serialization of the data between two systems. The first publication of the format was by the world wide web (WWW) consortium in 1998. As a big data engineer, what are the chances of dealing with this format? They are pretty good since industries such as health care, travel agencies, financial products, and mortgages use this standard to transfer data. How can we read and write XML files using Apache Spark?
Solution
Azure Databricks has provided the big data engineer with a library that can be used to work with XML files.
Business Problem
Our company has just started using Azure Databricks, and our manager wants us to learn how to manage XML files with Apache Spark. Here is a list of tasks that we need to investigate and solve:
| Task Id | Description |
|---|---|
| 1 | Sample XML Files |
| 2 | Configure Spark Cluster |
| 3 | Read XML Files |
| 4 | Write XML Files |
| 5 | Comparing File Formats |
At the end of the research, we will have a good understanding of how to manage XML files.
Sample Data Sets
I found three sample XML files on the internet that you can download. First, the book catalog dataset can be found on the Microsoft website.

Second, the compact disc (CD) catalog can be found on the XML Files website.

Third, the multiple record set XML file contains data on books, foods, and plants. This file can be downloaded from the Learning Container website.

The code will read these three XML files into a dataframe. Please download the files to your PC and upload them to the bronze zone of your data lake. The image below shows a listing of the Spark tips directory that contains the files:

As for writing XML files, we will use the airline data already stored in the hive catalog. My previous article had three managed tables related to airlines, airplanes, and airports. The image below shows the hive catalog with these three tables:
Now that we have our datasets, how do we configure our Spark cluster to work with XML files?
Configure Spark Cluster
Please edit the cluster named "interactive cluster," which currently has three libraries installed. Click the "Install new" button to start adding a new library.

It is very important to research the libraries you want to use. There can be compatibility issues when mixing a library built for the wrong Spark or Scala version. Use the following link to find details on the Databricks Spark driver for XML.

The image below shows the error I received when I installed the wrong library with Azure Databricks Runtime Engine (DBR). The below Scala class is not found. This issue can be resolved by uninstalling the non-compatible library and installing one compatible with the DBR.

The search feature for the Maven library is sometimes finicky. I just entered the fully qualified packaged name "com.databricks:spark-xml_2.12:0.16.0" to install the library.

After the installation, we can see that the new library is available on our cluster. Sometimes the cluster needs to be restarted for the library to become available.
Now that the cluster is configured, we can read in our first XML file.
Read XML File (Spark Dataframes)
The Spark library for reading XML has simple options. We must define the format as XML. We can use the rootTag and rowTag options to slice out data from the file. This is handy when the file has multiple record types. Last, we use the load method to complete the action. The code below reads the books catalog from our data lake.
#
# 1 - read in books
#
# set path to file
path1 = 'dbfs:/mnt/datalake/bronze/sparktips/book-catalog.xml'
# pick root and row tag
df1 = spark.read \
.format("xml") \
.option("rootTag", "catalog") \
.option("rowTag", "book") \
.load(path1)
# display the data
display(df1)
The output below shows the dataframe for the book catalog.

When dealing with text-based files, it is always best to supply a schema. That way, the Spark engine does not infer the data types. The code below uses the schema method of the read command to define the structure of the file. Note: The root and row tags are case-sensitive!
#
# 2 - read in cds
#
# set path to file
path2 = 'dbfs:/mnt/datalake/bronze/sparktips/cd-catalog.xml'
# create a schema
schema = 'ARTIST string, COMPANY string, COUNTRY string, PRICE double, TITLE string, YEAR long'
# pick root and row tag
df2 = spark.read \
.format("xml") \
.schema(schema) \
.option("rootTag", "CATALOG") \
.option("rowTag", "CD") \
.load(path2)
# display the data
display(df2)
The image below shows the results of displaying the CD catalog dataframe.

We can see that reading XML files into a Spark dataframe is quite easy.
Read XML File (Spark SQL)
As a Business Analyst, we might want to create a table in the hive catalog to analyze the data. How can we expose the previous XML files using Spark SQL syntax?
The code below uses the CREATE TABLE statement to achieve the desired result.
%sql -- -- 3 - Create table w/o schema -- CREATE TABLE sparktips.umt_catalog_of_books USING xml OPTIONS (path 'dbfs:/mnt/datalake/bronze/sparktips/book-catalog.xml', rowTag "book")
One can imagine that a SELECT statement would generate the same output as the display command used with the book catalog dataset. Let's do some analysis of the book catalog data. The following Spark SQL statement will find the max, min, and avg price of a book by genre.
%sql select genre, ROUND(MAX(PRICE), 2) as max_sale, ROUND(MIN(PRICE), 2) as min_sale, ROUND(AVG(PRICE), 2) as avg_sale from sparktips.umt_catalog_of_books group by genre
The image below shows five different book genres in the data file.

If I have not driven home the point yet, please use a schema when working with text-based files. This will improve the performance of loading the file into memory. The code below reads in the CD catalog file using a defined schema.
%sql -- -- 4 - Create table w schema -- CREATE TABLE sparktips.umt_catalog_of_cds ( ARTIST string, COMPANY string, COUNTRY string, PRICE double, TITLE string, YEAR long ) USING xml OPTIONS (path 'dbfs:/mnt/datalake/bronze/sparktips/cd-catalog.xml', rowTag "CD")
To make things interesting, let's find out if CD sales have different prices in different countries.
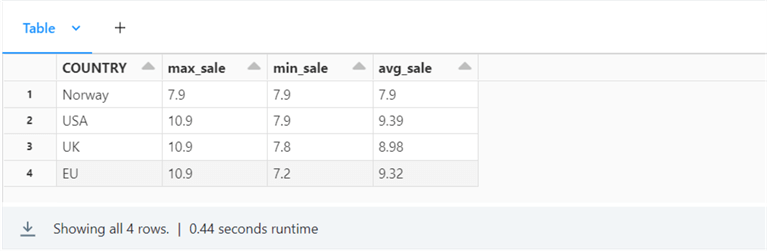
%sql select COUNTRY, ROUND(MAX(PRICE), 2) as max_sale, ROUND(MIN(PRICE), 2) as min_sale, ROUND(AVG(PRICE), 2) as avg_sale from sparktips.umt_catalog_of_cds group by COUNTRY
The image below shows the max, min, and average sale price of a CD in four different countries.
To recap, the Spark SQL syntax achieves the same result as the dataframe syntax. The only difference is that the file is exposed as a table in the hive catalog.
Read XML Files (Multiple Records)
I am going to use dataframes to read in the XML files. The code below reads in the multiple record file without a record tag. It will not fail but will produce an empty dataframe.
#
# 5 - read in root - does not work, read in row - works fine
#
# set path to file
path3 = 'dbfs:/mnt/datalake/bronze/sparktips/multiple-records.xml'
# pick root and row tag
df3 = spark.read \
.format("xml") \
.option("rootTag", "CATALOG") \
.load(path3)
# display the data
df3.printSchema
The image below shows an empty dataframe.

The code below uses both the root and row tags to extract PLANT records from the XML file. There is no way to read in all three record types without calling the Spark function three times.
#
# 6 - read in plants
#
# set path to file
path4 = 'dbfs:/mnt/datalake/bronze/sparktips/multiple-records.xml'
# pick root and row tag
df4 = spark.read \
.format("xml") \
.option("rootTag", "CATALOG") \
.option("rowTag", "PLANT") \
.load(path4)
# display the data
display(df4)
The image below shows the four PLANT records in the multiple record XML file.

The code below uses both the root and row tags to extract FOOD records from the XML file.
#
# 7 - read in food
#
# set path to file
path4 = 'dbfs:/mnt/datalake/bronze/sparktips/multiple-records.xml'
# pick root and row tag
df4 = spark.read \
.format("xml") \
.option("rootTag", "CATALOG") \
.option("rowTag", "FOOD") \
.load(path4)
# display the data
display(df4)
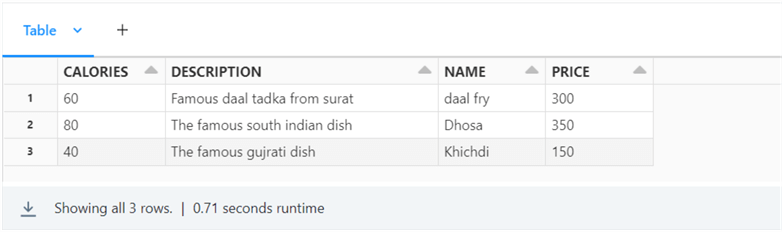
The image below shows the three FOOD records in the multiple record XML file.
The code below uses both the root and row tags to extract BOOK records from the XML file.
#
# 8 - read in book
#
# set path to file
path4 = 'dbfs:/mnt/datalake/bronze/sparktips/multiple-records.xml'
# pick root and row tag
df4 = spark.read \
.format("xml") \
.option("rootTag", "CATALOG") \
.option("rowTag", "BOOK") \
.load(path4)
# display the data
display(df4)
The image below shows the three BOOK records in the multiple record XML file.

In a nutshell, reading different record types from an XML file requires multiple file reads. If this was a real production problem, consider parameterizing a function that would take a list of row tags as input and create an array of dataframes as output. This will drastically reduce the code. Additionally, I would pass a list of schema definitions so the engine would perform at its best. Now that we have dealt with multiple ways to read an XML file, we will concentrate on writing a large amount of data to XML files.
Write XML File (Spark Dataframes)
Let's imagine that the Federal Aviation Administration (FAA) has asked our company to download all historical airline data by year to XML files. How can we write a set of Spark notebook cells to perform this action?
First, let's take a look at the data. We want to include only flight data with airplane tail numbers. The SELECT statement below returns the results we want.
%sql select * from sparktips.mt_airline_data where TailNum is not null
By default, the Azure Databricks control plane or GUI only returns the first 1000 rows.

We can write a simple aggregation query to find the number of years in the hive table and how many records per year:
%sql select FltYear, count(*) as Flttotal from sparktips.mt_airline_data where TailNum is not null group by FltYear order by FltYear
We can see from the image below that the table has 22 years of data. Also, if we scan the data, we can see the row count is between 5-8 million rows for a complete year of data collection.

I have shared the toolbox notebook, which has code to take a single partitioned spark file and create a single resulting file. This code needs to be modified to work with the XML file output from the spark.write command. Unlike other file formats, a file extension is not given. The code snippet shows the update. If we do not pass a file extension, look for any file in the output directory starting with the "part*" file pattern.
# find new file
if file_ext == "":
tmp_lst = get_file_list(tmp_dir, "part*")
else:
tmp_lst = get_file_list(tmp_dir, "part*." + file_ext)
To include this update toolbox code, use the %run magic command to add the functions to the current spark session.
%run "./nb-tool-box-code"
While the code for this multiple file write is large, the algorithm is straightforward. For each year, create an XML file of airline data:
| Task Id | Description |
|---|---|
| 1 | Get a list of airline years |
| 2A | For each year, write out a temporary XML directory |
| 2B | For each XML directory, move XML file to final directory and file name |
| 2C | Remove the temporary XML directory |
Note: This code is repeatable. It removes the audit directory before creating the XML files. It is very important to repartition the dataframe so that only 1 XML file is generated per year. The unwanted_file_cleanup() function handles the file movement with the temporary Spark directory that contains the 1 XML file.
#
# 9 - write xml files by year for flight data
#
# del old dir
try:
dbutils.fs.rmdir("dbfs:/mnt/datalake/bronze/sparktips/audit/")
except:
pass
# make new dir
dbutils.fs.mkdirs("dbfs:/mnt/datalake/bronze/sparktips/audit/")
# sql - get a list of years
sql_stmt1 = """
select distinct FltYear
from sparktips.mt_airline_data
order by FltYear
"""
# df - get a list of years
df_years = spark.sql(sql_stmt1)
# for each year
for row_year in df_years.rdd.collect():
# show progress
msg = "processing data for year {}".format(row_year[0])
print(msg)
# sql - get flight data w/ tail nums
sql_stmt2 = """
select *
from sparktips.mt_airline_data
where TailNum is not null and FltYear = {}
""".format(row_year[0])
# df - get flight data w/ tail nums
df_airline = spark.sql(sql_stmt2)
# set path
path1 = "/mnt/datalake/bronze/sparktips/temp"
path2 = "/mnt/datalake/bronze/sparktips/audit/flights-year-{}.xml".format(row_year[0])
# write xml file
df_airline.repartition(1).write \
.format("xml") \
.option("rootTag", "flights") \
.option("rowTag", "flight") \
.save(path1)
# create single file
unwanted_file_cleanup(path1, path2, "")
Since a large amount of data is being created, I included a print statement to see the progress of the program. Two datasets are being used. The first one has the number of years of airline data. The second has the actual airline data for a given year.

The output from the above Python code can be found in the final section of this article.
Comparing File Formats
We want to write some code that will help us gather file sizes for directories that might be nested. That way, we can compare the original size of the airline data (parquet format) to the current size (XML files) requested for the fictitious FAA audit.
The code below uses the file system magic command (%fs) to list the files in the audit directory. This command does not work well with nested directories.
%fs ls /mnt/datalake/bronze/sparktips/audit
The output shows two plus gigabyte files in the audit directory. There are no sub-directories which makes this easy to summarize.

We can use the Azure Portal to validate the file sizes also.

If we download the file to our local personal computer, we can open it up in Notepad++. We can see that the data file for 1987 has 22.3 M records.

We can use the os and subprocess libraries to write a shell function that returns the execution output as a text string:
#
# 10 - create function to capture output
#
# import libraries
from os import getcwd
from subprocess import check_output
# return output from shell
def sh(command):
return check_output(command, shell=True, cwd=getcwd(), universal_newlines=True).strip()
The du Linux command will return the desired results, showing size totals per directory. The image below saves the text output of the parquet files in a variable called output1.
#
# 11 - show parquet files (hive tables)
#
# get size of parquet files
output1 = sh("du -h /dbfs/user/hive/warehouse/sparktips.db/mt_airline_data/")
print(output1)
There are over 200 file directories in the hive catalog since we partitioned on year and month for the 22 years. The output below is a huge list in a text variable. How can we sum up this data to get the final size of all files?

The image below saves the text output of the XML files in a variable called output2.
#
# 12 – show xml files (requested audit)
#
output2 = sh("du -h /dbfs/mnt/datalake/bronze/sparktips/audit")
print(output2)
The output from the audit directory gives us an answer of 55 G for the total size. But this is not in actual bytes.

The code below defines a function called parse_output_n_sum_size. It takes the string output of the du command as input. Both the pandas and io libraries are used to solve the problem. The output is the actual number of bytes calculated by mapping the output from the du command to numeric values.
#
# 13 - write functions to parse + sum file sizes
#
import pandas as pd
from io import StringIO
# convert alpha to numeric
def size_to_bytes(a):
if a == "K":
return 1000
elif a == "M":
return 1000000
elif a == "G":
return 1000000000
elif a == "T":
return 1000000000000
else:
return 1
# get total bytes
def parse_output_n_sum_size(input1):
# string io
data1 = StringIO(input1)
# covert to pandas
df = pd.read_csv(data1, sep ="\t", names=["size", "path"])
# grab type + size
df['type'] = df['size'].str[-1:]
df['value'] = df['size'].str[:-1]
# map to bytes + calculate
df['bytes'] = df['type'].apply(size_to_bytes)
df['size_in_bytes'] = df.value.astype(float) * df.bytes.astype(int)
# return total size
total = df['size_in_bytes'].sum()
return total
Let's go into more detail about the above code. The StringIO function converts the string into a byte stream. Pandas has the read_csv function that converts the byte stream into a dataframe with a header. String slicing is used to split the size value from the size indicator. Thus, a string with the value of "2.9M" needs to be split into two pieces. The apply method maps the indicator into bytes using a custom function called size_to_bytes. Thus, the M, for mega-bytes, is turned into 1 million logical bytes. The astype method is used to cast a column in a Pandas dataframe to a different base datatype such as float. To finish the coding, the sum method is used to return the total bytes of the Pandas dataframe.
The image below shows three cells. The first one calculates the total bytes of the partition hive table, which is around 1.98 GB. The second one calculates the total bytes of the XML files, which is about 55 GB. As we know, text formats such as XML and JSON expand the data size due to repeated tagging. Thus, the XML file format takes up 27.8 times more space than the parquet format.

One can conclude that Python libraries are useful for management tasks. It would have been difficult to write a Spark program using the dbutils.fs.ls command to recursively find all the files in the partitioned hive delta table. However, it was easy to use Python libraries to figure out the answer. I find that I use the Panda's library to solve problems that Spark dataframes do not support.
Next Steps
Using Apache Spark to read and write XML data files can be easily done. The record tag is the only option that is required. However, it is best practice to supply both the root and record tags, which are case-sensitive. Spark supplies the big data engineer with syntax to read the XML file into a dataframe or manage the file as a hive table. In both cases, it is a best practice to supply the file schema to increase performance. The one issue that you might encounter with an XML file is the storage of multiple record types in one file. In this rare case, you will have to read the XML file multiple times to extract each record type. In short, working with XML files is quite easy.
Is XML an efficient file format to use with Apache Spark? The answer is no. We saw that the XML file format was more than 25 times larger than the delta file format, which uses parquet files. As a big data developer, will you be asked to work with XML files? The answer is yes. Given the number of industries that use this format for data exchange, you will more than likely be asked to process this file format in the future.
Enclosed are the zip files that contain the following information: the data files zip and the Spark code zip.
About the author
 John Miner is a Data Architect at Insight Digital Innovation helping corporations solve their business needs with various data platform solutions.
John Miner is a Data Architect at Insight Digital Innovation helping corporations solve their business needs with various data platform solutions.This author pledges the content of this article is based on professional experience and not AI generated.
View all my tips
Article Last Updated: 2023-03-21






